
“A few weeks is what most clinicians call ‘rapid’ when it comes to sequencing a patient’s genome and returning results.”
Dr. Euan Ashley, professor of medicine, genetics and biomedical data science, Stanford School of Medicine
In a new milestone, Dr. Euan Ashley and his colleagues Stanford School of Medicine used AI to sequence and analyze the genome of a hospitalized patient in 5 hours and 2 minutes. This is the fastest DNA sequencing technique ever developed for genetic diagnostics. These researchers hold the record for the fastest genetic diagnosis in a critical care setting – 7 hours 18 minutes! For comparison, traditional genetic sequencing and testing takes weeks. The record was certified by the National Institute of Science and Technology’s Genome in a Bottle group and is documented by Guinness World Records.
Genome sequencing allows scientists to see a patient’s complete DNA makeup including information about inherited diseases. Accelerating genetic diagnosis is important because it can help doctors diagnose rare genetic diseases. Once doctors know the specific genetic mutation, they can tailor treatments in a very precise way. This new method can guide clinical management, improve prognosis, and reduce costs for patients. The ultrarapid technique accelerated every step of genome sequencing workflow and resulted in rapid and reliable results.
One of the patients in the study was a 3-month-old full-term infant who was having epileptic seizures. Magnetic resonance imaging of the brain revealed no abnormalities. The researchers used the new method and in 8 hours and 25 minutes they identified a likely pathogenic heterozygous variant in CSNK2B. This variant and gene are known to cause a neurodevelopmental disorder with early-onset epilepsy. The researchers made a definitive diagnosis of a CSNK2B-related disorder called Poirier–Bienvenu neurodevelopmental syndrome. Since they diagnosed the disorder in 8 hours, the baby did not have to undergo additional diagnostic testing and the doctors were able to begin treatment.
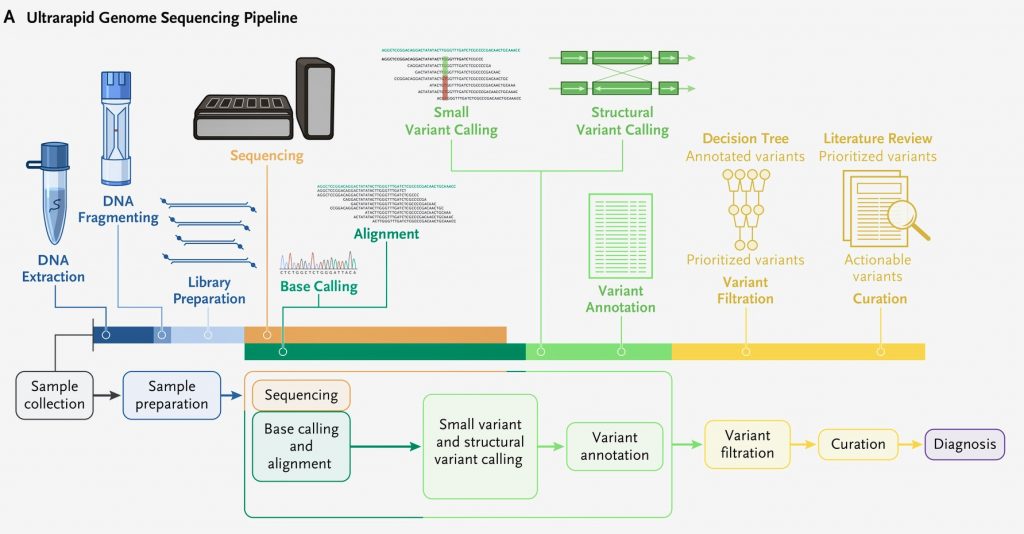
Study Overview
The study entitled Ultrarapid Nanopore Genome Sequencing in a Critical Care Setting was published in the New England Journal of Medicine. The research was led by Dr Euan Ashley, professor of medicine, genetics and biomedical data science at Stanford School of Medicine. Dr. Euan and his team collaborated with NVIDIA, Oxford Nanopore Technologies, Google, Baylor College of Medicine, and the University of California. The authors described the method they used to rapidly sequence and analyze of the genomes of twelve patients. Five of the patients in the study received a diagnosis.
“I think we can halve it again. If we’re able to do that, we’re talking about being able to get an answer before the end of a hospital ward round. That’s a dramatic jump.”
Dr. Euan Ashley, professor of medicine, genetics and biomedical data science, Stanford School of Medicine
Ultra-fast Sequencing
To achieve super-fast sequencing speeds the researchers needed new hardware. The researchers used nanopore sequencing on Oxford Nanopore’s PromethION Flow Cells to generate more than 100 gigabases of data per hour, and NVIDIA GPUs on Google Cloud to speed up the base calling and variant calling processes. Genomic data overwhelmed the lab’s computational systems and they weren’t able to process the data fast enough. They had to completely rethink their data pipelines and storage systems.
Graduate student Sneha Goenka found a way to funnel the data straight to a cloud-based storage system where computational power could be amplified enough to sift through the data in real time and the Stanford Research Computing Center advised the team on the network and bandwidth requirements needed to complete the project. AI algorithms scanned the incoming genetic code for errors that might cause disease and compared the patients’ gene variants against publicly documented variants known to cause disease. Oxford Nanopore Technologies contributed to the cost of sequencing and reagents, Google contributed to the cost of cloud computing, and NVIDIA contributed to the cost of Parabricks. This is an excellent example of the application of high performance computing to healthcare.
Cost Savings
The estimated costs of this approach including DNA extraction, library preparation, sequencing, and computation range from $4,971 – $7,318. Since the daily cost of critical care is more than $10,000, rapid genome sequencing diagnostics can significantly reduce costs. Because of the significant cost savings, rapid genome sequencing diagnostics has been reimbursed by several insurance companies.

Study Highlights
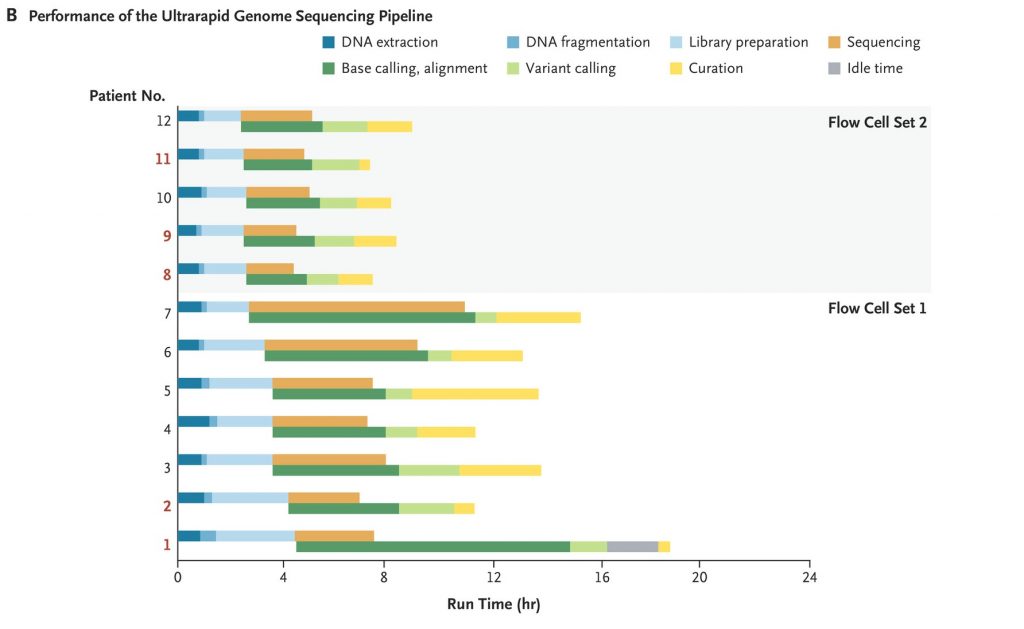
The study was conducted at two hospitals in Stanford California between December 2020 and May 2021. In less than six months the team enrolled and sequenced the genomes of 12 patients. The team’s diagnostic rate was 42% which is 12% higher than the average rate for diagnosing mystery diseases.
- Researchers enrolled 12 patients who were generally representative of persons living in the United States with respect to race, ethnic group, and sex.
- Researchers obtained an initial genetic diagnosis in 5 of the patients. The shortest time from arrival of the blood sample in the laboratory to the initial diagnosis was 7 hours 18 minutes.
- After establishing a diagnosis in Patient 1, they updated their bioinformatics framework to permit the transfer of terabytes of raw signal data to Cloud storage in real time and distributed the data across multiple Cloud computing machines to achieve near real-time base calling and alignment. This step reduced the postsequencing run time by 93%, from 7 hours 21 minutes to 34 minutes.
- Flow cells were washed and reused until exhaustion to reduce the sequencing cost per sample.
- Libraries were bar-coded in Patients 1 through 7 to prevent carryover from one sample to the next.
- After processing the sample obtained from Patient 7, they benchmarked and adopted a bar-code–free method to rapidly generate genome sequences. Removing the bar-coding process accelerated sample preparation by 37 minutes, to an average of 2.5 hours, and enabled them to load a greater amount of patients’ DNA into each flow cell (333 ng vs. 155 ng) and increase pore occupancy (to 82% from 64%)
- Their sequencing workflow generated 173 to 236 Gb of data per genome using 48 flow cells, with an alignment identity of 94% and 46 to 64× autosomal coverage. Half the sequencing throughput was in reads that were 25 kb or longer.
- Small variants and structural variants called after the reads were aligned to the GRCh37 human reference genome, which generated a median of 4,490,490 single-nucleotide variants and small insertions and deletions.
- Custom filtration and prioritization of variants with an ultrarapid scoring system substantially decreased the number of candidate variants for manual review to a median of 29 (range 16 to 53) for small variants and 22 (range 11 to 37) for structural variants
- Each initial diagnosis was immediately reviewed by study and bedside physicians, and a consensus was reached as to whether the proposed variant represented the primary cause of the patient’s presentation.
- Diagnostic variants were identified in 5 of the 12 patients, who ranged in age from 3 months to 57 years.
- The findings were immediately confirmed by a laboratory certified by the Clinical Laboratory Improvement Amendments process and informed clinical management including sympathectomy, heart transplantation, screening, and changes in medication, for each of the 5 patients or their family members.
Dr. Euan Ashley

Dr. Ashley was born in Scotland and graduated with 1st class Honors in Physiology and Medicine from the University of Glasgow. He completed medical residency and a PhD at the University of Oxford before moving to Stanford University where he trained in cardiology and advanced heart failure, joining the faculty in 2006. His group is focused on the science of precision medicine. In 2010, Dr. Ashley led the team that carried out the first clinical interpretation of a human genome. The article became one of the most cited in clinical medicine that year and was later featured in the Genome Exhibition at the Smithsonian in DC. Over the following 3 years, the team extended the approach to the first whole genome molecular autopsy, to a family of four, and to a case series of patients in primary care. They now routinely apply genome sequencing to the diagnosis of patients at Stanford hospital where Dr Ashley directs the Clinical Genome Program and the Center for Inherited Cardiovascular Disease.

In 2021, his first book The Genome Odyssey – Medical Mysteries and the Incredible Quest to Solve Them was released. Dr Ashley has a passion for rare genetic disease and was the first co-chair of the steering committee of the Undiagnosed Diseases Network. He was a recipient of the National Innovation Award from the American Heart Association and the NIH Director’s New Innovator Award. He is part of the winning team of the $75m One Brave Idea competition and co-founder of three companies: Personalis Inc ($PSNL), Deepcell Inc, and SVExa Inc. He was recognized by the Obama White House for his contributions to Personalized Medicine and in 2018 was awarded the American Heart Association Medal of Honor for Genomic and Precision Medicine. He was appointed Stanford Associate Dean in 2019.
